This package is a suite of helper functions that can be used at Gavi. It has functions to help simplify repeat procedures, like loading common data, gavi specific colors and themes for plots and tables, and functions which add and filter common varibles like country groupings and iso3 codes.
Installation
You can install the development version of gavir from GitHub with:
# install.packages("devtools")
devtools::install_github("joshualorin/gavir")Examples
You can use the get_*() functions to load data:
library(gavir)
library(tidyverse, warn.conflicts = F, verbose = F)
#> ── Attaching packages ─────────────────────────────────────── tidyverse 1.3.2 ──
#> ✔ ggplot2 3.4.3 ✔ purrr 1.0.2
#> ✔ tibble 3.2.1 ✔ dplyr 1.1.2
#> ✔ tidyr 1.3.0 ✔ stringr 1.5.0
#> ✔ readr 2.1.4 ✔ forcats 0.5.2
#> ── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
#> ✖ dplyr::filter() masks stats::filter()
#> ✖ dplyr::lag() masks stats::lag()
# all functions require a version date
get_groupings("2023-07")
#> Reading in groupings file dated: 2023-07
#> Interesting! The groupings file you have selected is not the most recent. If this was unintentional, you can update to the most recent file, which is 2023-08
#> # A tibble: 196 × 61
#> iso3 iso3_num iso2 country country_short lang global gavi77 gavi73 gavi72
#> <chr> <dbl> <chr> <chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 AFG 4 AF Afghani… Afghanistan eng 1 1 1 1
#> 2 AGO 24 AO Angola Angola eng 1 1 1 1
#> 3 ALB 8 AL Albania Albania eng 1 1 0 0
#> 4 AND 20 AD Andorra Andorra eng 1 0 0 0
#> 5 ARE 784 AE United … United Arab … eng 1 0 0 0
#> 6 ARG 32 AR Argenti… Argentina eng 1 0 0 0
#> 7 ARM 51 AM Armenia Armenia eng 1 1 1 1
#> 8 ATG 28 AG Antigua… Antigua and … eng 1 0 0 0
#> 9 AUS 36 AU Austral… Australia eng 1 0 0 0
#> 10 AUT 40 AT Austria Austria eng 1 0 0 0
#> # ℹ 186 more rows
#> # ℹ 51 more variables: gavi68 <dbl>, gavi57 <dbl>, gavi55 <dbl>, mics45 <dbl>,
#> # dov96 <dbl>, vimc112 <dbl>, who_region <chr>, gavi_region <chr>,
#> # gavi_region_short <chr>, pef_type <chr>, continental_africa <dbl>,
#> # francophone <chr>, indo_pacific <dbl>, regional_je <dbl>,
#> # regional_mena <dbl>, regional_yfv_2016 <dbl>, regional_yfv_2020 <dbl>,
#> # regional_ipv <dbl>, segments_2020 <chr>, segments_2021 <chr>, …
# and most have additional parameters you can fiddle with
get_ihme("2023-02", source = "subnational", vaccine = "dtp3")
#> Reading in Subnational IHME file dated 2023-02 and aggregated to polio shapes
#> Rows: 1217964 Columns: 17── Column specification ────────────────────────────────────────────────────────
#> Delimiter: ","
#> chr (8): iso3, admin2, admin2_code, vaccine, admin1, admin1_code, pop_model,...
#> dbl (9): year, coverage, pu1_final, unvax_total, pct_unvax, upper, lower, pu...
#> ℹ Use `spec()` to retrieve the full column specification for this data.
#> ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
#> # A tibble: 405,988 × 17
#> iso3 admin2 admin2_code year vaccine coverage pu1_final unvax_total
#> <chr> <chr> <chr> <dbl> <chr> <dbl> <dbl> <dbl>
#> 1 ETH MAO KOMO SPEC… {00061F00-… 2000 dtp3 0.174 1723. 1424.
#> 2 ETH MAO KOMO SPEC… {00061F00-… 2001 dtp3 0.269 1760. 1287.
#> 3 ETH MAO KOMO SPEC… {00061F00-… 2002 dtp3 0.255 1792. 1335.
#> 4 ETH MAO KOMO SPEC… {00061F00-… 2003 dtp3 0.215 1807. 1419.
#> 5 ETH MAO KOMO SPEC… {00061F00-… 2004 dtp3 0.259 1846. 1368.
#> 6 ETH MAO KOMO SPEC… {00061F00-… 2005 dtp3 0.299 1885. 1320.
#> 7 ETH MAO KOMO SPEC… {00061F00-… 2006 dtp3 0.339 1906. 1261.
#> 8 ETH MAO KOMO SPEC… {00061F00-… 2007 dtp3 0.385 1931. 1188.
#> 9 ETH MAO KOMO SPEC… {00061F00-… 2008 dtp3 0.455 1955. 1065.
#> 10 ETH MAO KOMO SPEC… {00061F00-… 2009 dtp3 0.485 1958. 1007.
#> # ℹ 405,978 more rows
#> # ℹ 9 more variables: pct_unvax <dbl>, admin1 <chr>, admin1_code <chr>,
#> # pop_model <chr>, pop_adjustment <chr>, upper <dbl>, lower <dbl>,
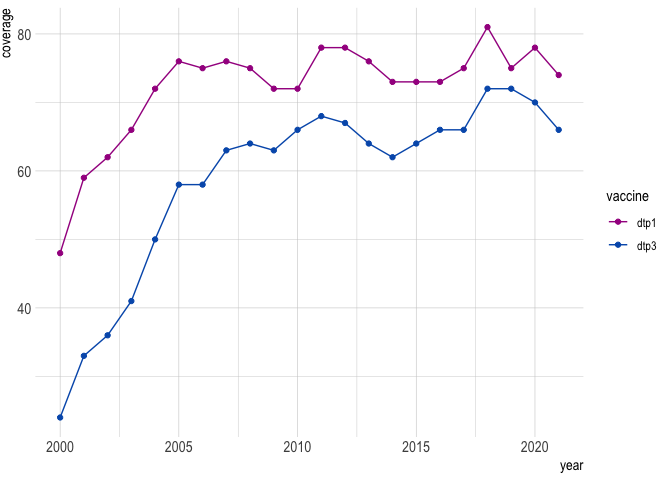
#> # pu1_uncalib <dbl>, pu1_calib <dbl>theme_*() and scale_*() functions allow for standard gavi formatting and colors:
wuenic %>%
filter(iso3 == "AFG" & vaccine %in% c("dtp1", "dtp3")) %>%
ggplot(aes(year, coverage, color = vaccine)) +
geom_line() +
geom_point() +
scale_color_gavi() +
theme_gavi()